Functionality 1: detect dubious metacells for a given metacell partition
Pan Liu
18 October 2025
mcRigor-1-detect-dubmc.RmdIntroduction
In this tutorial, we will show how to use mcRigor to detect dubious metacells for a given metacell partition. We will demonstrate this functionality of mcRigor on a semi-synthetic single cell RNA sequencing (scRNA-seq) dataset with known ground truth trustworthiness of metacells.
Input preparation
Two main inputs are required for this functionality: 1. the raw
scRNA-seq data and 2. a given metacell partition generated by either
existing metacell partitioning methods or ad hoc approaches. The raw
scRNA-seq data needs to be provided as a Seurat object,
obj_singlecell. The semi-synthetic scRNA-seq data, whose
generation process is described in Liu
and Li, 2024, stored as a rds file syn.rds, is
available with the mcRigor package as an example. We first load the
data.
sc_dir = system.file('extdata', 'syn.rds', package = 'mcRigor')
obj_singlecell= readRDS(file = sc_dir)
obj_singlecell
#> An object of class Seurat
#> 2000 features across 13400 samples within 1 assay
#> Active assay: RNA (2000 features, 2000 variable features)
#> 3 layers present: counts, data, scale.data
#> 2 dimensional reductions calculated: pca, umapThe metacell partition should be provided as a dataframe,
cell_membership, showing the assignment of single cells to
metacells. Specifically, this dataframe has only one column and each row
of the dataframe represents one single cell. The metacell partitions for
the semi-synthetic scRNA-seq data generated by the SEACells method (Persad et al.,
2023), stored as a csv file
seacells_cell_membership_rna_syn.csv, is available with the
mcRigor package as an example. This csv file contains series of metacell
partitions, which were generated under different granularity levels.
Note that granularity level,
,
is a key parameter for metacell partitioning and is defined as the ratio
of the number of single cells to the number of metacells. In this
tutorial, we focus on the metacell partition given by granularity level
.
membership_dir = system.file('extdata', 'seacells_cell_membership_rna_syn.csv', package = 'mcRigor')
cell_membership_all <- read.csv(file = membership_dir, check.names = F, row.names = 1)
cell_membership <- cell_membership_all['50']
head(cell_membership)
#> 50
#> 1_Cell1 mc50-allcells-SEACell-98
#> 2_Cell1 mc50-allcells-SEACell-98
#> 3_Cell1 mc50-allcells-SEACell-98
#> 4_Cell1 mc50-allcells-SEACell-98
#> 5_Cell1 mc50-allcells-SEACell-98
#> 6_Cell1 mc50-allcells-SEACell-98Detection of dubious metacells
We call the function mcRigor_DETECT to detect dubious
metacells for the metacell partition represented by
cell_membership.
detect_res = mcRigor_DETECT(obj_singlecell = obj_singlecell, cell_membership = cell_membership)Alternatively, we can input all metacell partitions
(cell_membership_all) into the function
mcRigor_DETECT and specify the partition of interest by
setting the argument tgamma. The code below indicates that
we aim to detect dubious metacells for the partition under granularity
level tgamma = 50, while borrowing information from other
metacell partitions during null construction. This approach is
recommended when additional computation time is acceptable, as it
generally produces more stable results.
detect_res = mcRigor_DETECT(obj_singlecell = obj_singlecell, cell_membership = cell_membership_all, tgamma = 50)The Seurat object of metacells are stored in the
obj_metacell field of the output
detect_res.The mcRigor detection results are recorded in
the mc_res field of the output detect_res as
well as the metadata of the Seurat object with name
mcRigor.
table(detect_res$mc_res)
#>
#> dubious trustworthy
#> 56 212
obj_metacell = detect_res$obj_metacell
head(obj_metacell$mcRigor)
#> mc50-allcells-SEACell-0 mc50-allcells-SEACell-1 mc50-allcells-SEACell-10
#> "trustworthy" "dubious" "dubious"
#> mc50-allcells-SEACell-100 mc50-allcells-SEACell-101 mc50-allcells-SEACell-102
#> "trustworthy" "trustworthy" "trustworthy"Visualization
The function mcRigor_projection can draw the metacells
projected to the two-dimensional embedding space of single cells and
mark the detected dubious metacells
sc_membership = obj_metacell@misc$cell_membership$Metacell
names(sc_membership) = rownames(obj_metacell@misc$cell_membership)
plot = mcRigor_projection(obj_singlecell = obj_singlecell, sc_membership = sc_membership,
color_field = 'celltype.l1',
dub_mc_test.label = T, test_stats = detect_res$TabMC, Thre = detect_res$thre)
plot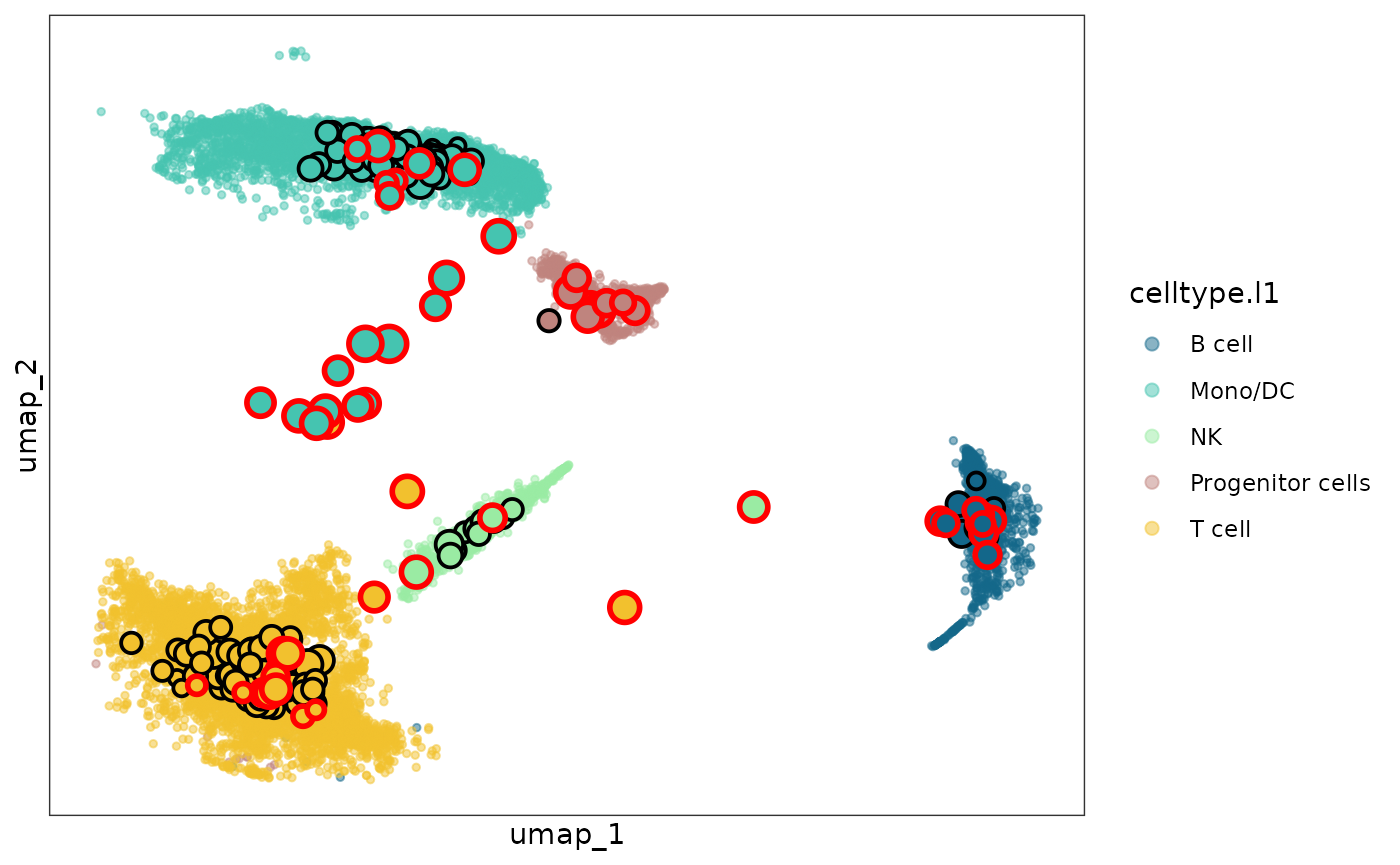
The dubious metacells are marked by red circles while the trustworthy metacells are with black circles.
Session information
sessionInfo()
#> R version 4.5.1 (2025-06-13)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 24.04.3 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] ggplot2_4.0.0 Seurat_5.3.0 SeuratObject_5.2.0 sp_2.2-0
#> [5] mcRigor_1.0 BiocStyle_2.36.0
#>
#> loaded via a namespace (and not attached):
#> [1] deldir_2.0-4 pbapply_1.7-4 gridExtra_2.3
#> [4] rlang_1.1.6 magrittr_2.0.4 RcppAnnoy_0.0.22
#> [7] spatstat.geom_3.6-0 matrixStats_1.5.0 ggridges_0.5.7
#> [10] compiler_4.5.1 reshape2_1.4.4 png_0.1-8
#> [13] systemfonts_1.3.1 vctrs_0.6.5 stringr_1.5.2
#> [16] pkgconfig_2.0.3 fastmap_1.2.0 labeling_0.4.3
#> [19] promises_1.3.3 rmarkdown_2.30 ragg_1.5.0
#> [22] purrr_1.1.0 xfun_0.53 cachem_1.1.0
#> [25] jsonlite_2.0.0 goftest_1.2-3 later_1.4.4
#> [28] spatstat.utils_3.2-0 irlba_2.3.5.1 parallel_4.5.1
#> [31] cluster_2.1.8.1 R6_2.6.1 ica_1.0-3
#> [34] spatstat.data_3.1-8 stringi_1.8.7 bslib_0.9.0
#> [37] RColorBrewer_1.1-3 reticulate_1.43.0 spatstat.univar_3.1-4
#> [40] parallelly_1.45.1 lmtest_0.9-40 jquerylib_0.1.4
#> [43] scattermore_1.2 Rcpp_1.1.0 bookdown_0.45
#> [46] knitr_1.50 tensor_1.5.1 future.apply_1.20.0
#> [49] zoo_1.8-14 sctransform_0.4.2 httpuv_1.6.16
#> [52] Matrix_1.7-3 splines_4.5.1 igraph_2.2.0
#> [55] tidyselect_1.2.1 abind_1.4-8 yaml_2.3.10
#> [58] spatstat.random_3.4-2 spatstat.explore_3.5-3 codetools_0.2-20
#> [61] miniUI_0.1.2 listenv_0.9.1 plyr_1.8.9
#> [64] lattice_0.22-7 tibble_3.3.0 withr_3.0.2
#> [67] shiny_1.11.1 S7_0.2.0 ROCR_1.0-11
#> [70] evaluate_1.0.5 Rtsne_0.17 future_1.67.0
#> [73] fastDummies_1.7.5 desc_1.4.3 survival_3.8-3
#> [76] polyclip_1.10-7 fitdistrplus_1.2-4 pillar_1.11.1
#> [79] BiocManager_1.30.26 KernSmooth_2.23-26 plotly_4.11.0
#> [82] generics_0.1.4 RcppHNSW_0.6.0 scales_1.4.0
#> [85] globals_0.18.0 xtable_1.8-4 glue_1.8.0
#> [88] lazyeval_0.2.2 tools_4.5.1 data.table_1.17.8
#> [91] RSpectra_0.16-2 RANN_2.6.2 fs_1.6.6
#> [94] dotCall64_1.2 cowplot_1.2.0 grid_4.5.1
#> [97] tidyr_1.3.1 nlme_3.1-168 patchwork_1.3.2
#> [100] cli_3.6.5 spatstat.sparse_3.1-0 textshaping_1.0.4
#> [103] spam_2.11-1 viridisLite_0.4.2 dplyr_1.1.4
#> [106] uwot_0.2.3 gtable_0.3.6 sass_0.4.10
#> [109] digest_0.6.37 progressr_0.17.0 ggrepel_0.9.6
#> [112] htmlwidgets_1.6.4 farver_2.1.2 htmltools_0.5.8.1
#> [115] pkgdown_2.1.3 lifecycle_1.0.4 httr_1.4.7
#> [118] mime_0.13 MASS_7.3-65