Evaluate pseudotime goodness-of-fit
Introduction
In this example, we will show how to use scDesign3Py to evaluate the pseudotime goodness-of-fit for different pseudotime labels. If the true labels are unavailable and we have little prior knowledge, the scDesign3 BIC can serve as an unsupervised metric. In this tutorial, we will first use the ground truth “pseudotime” generated by the R package dyngen. Then, we will perturb the ground truth pseudotime to worsen its quality and use scDesign3’s BIC to examine pseudotime goodness-of-fit.
Import packages and Read in data
import pacakges
import anndata as ad
import numpy as np
import pandas as pd
import scDesign3Py
Read in data
The raw data is from the R package dyngen. Here, we directly use the result from the scDesign3 tutorial.
data = ad.read_h5ad("data/dyngen.h5ad")
data
AnnData object with n_obs × n_vars = 500 × 155
obs: 'step_ix', 'simulation_i', 'sim_time', 'pseudotime'
var: 'module_id', 'basal', 'burn', 'independence', 'color', 'is_tf', 'is_hk', 'transcription_rate', 'splicing_rate', 'translation_rate', 'mrna_halflife', 'protein_halflife', 'mrna_decay_rate', 'protein_decay_rate', 'max_premrna', 'max_mrna', 'max_protein', 'mol_premrna', 'mol_mrna', 'mol_protein'
uns: 'traj_dimred_segments', 'traj_milestone_network', 'traj_progressions'
obsm: 'dimred'
layers: 'counts_protein', 'counts_spliced', 'counts_unspliced', 'logcounts'
Simulation
We perturb the pseudotime by generating random numbers from uniform distribution and replacing various percentages of the original pseudotime with random numbers. The percentage ranges from 0% to 100%. In the code below, we generate 11 sets of perturbed pseudotime with the percentage of perturbation ranging from 0% to 100%. For each new set of perturbed pseudotime, we create a new AnnData object, storing the original count matrix and the corresponding perturbed pseudotime.
perturb_data_dict = {0:{"dat":data}}
for i in range(1,11):
tmp_data = data.copy()
num = round(i/10 * len(tmp_data))
np.random.seed(i)
tmp_data.obs.loc[tmp_data.obs.sample(n=num,random_state=i).index,"pseudotime"] = np.random.uniform(size=num)
perturb_data_dict[i] = {"dat":tmp_data}
Then we run scDesign3Py to get the model bic.
for key, value in perturb_data_dict.items():
tmp_data = value["dat"]
test = scDesign3Py.scDesign3(n_cores=2, parallelization="pbmcmapply")
test.set_r_random_seed(123)
res = test.scdesign3(anndata=tmp_data,
pseudotime="pseudotime",
corr_formula = "ind",
mu_formula = "s(pseudotime, bs = 'cr', k = 10)",
sigma_formula = "1",
copula = "gaussian",
default_assay_name = "counts",
family_use = "nb",
usebam = False)
perturb_data_dict[key]["bic"] = res["model_bic"]
Visualization
After the simulations, we use BIC, which is an unsupervised metric, for evaluating the goodness-of-fit of the pseudotime.
bic_res = pd.concat([i["bic"] for _,i in perturb_data_dict.items()],axis=1).T
bic_res.index = [f"{i/10} perturb" for i in bic_res.index]
bic_res
| bic.marginal | bic.copula | bic.total | |
|---|---|---|---|
| 0.0 perturb | 440744.638950 | 0.0 | 440744.638950 |
| 0.1 perturb | 465098.582392 | 0.0 | 465098.582392 |
| 0.2 perturb | 471527.503356 | 0.0 | 471527.503356 |
| 0.3 perturb | 487017.888671 | 0.0 | 487017.888671 |
| 0.4 perturb | 487621.784061 | 0.0 | 487621.784061 |
| 0.5 perturb | 493888.709002 | 0.0 | 493888.709002 |
| 0.6 perturb | 496339.699622 | 0.0 | 496339.699622 |
| 0.7 perturb | 499465.894561 | 0.0 | 499465.894561 |
| 0.8 perturb | 499858.520011 | 0.0 | 499858.520011 |
| 0.9 perturb | 501529.672888 | 0.0 | 501529.672888 |
| 1.0 perturb | 502280.653546 | 0.0 | 502280.653546 |
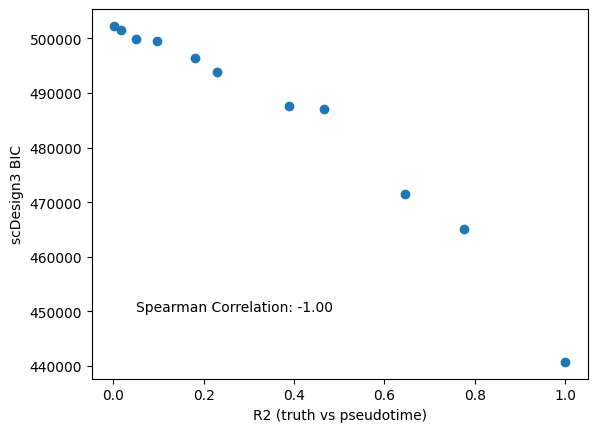
Since we also have the ground truth pseudotime, we also calculate the \(r^2\) between the ground truth pseudotime and perturbed pseudotime. The \(r^2\) is a supervised metric to evaluate the pseudotime qualities. The figure shows that model BICs agree with \(r^2\)
import matplotlib.pyplot as plt
r = []
for i in range(0,11):
r.append(perturb_data_dict[i]["dat"].obs["pseudotime"].corr(perturb_data_dict[0]["dat"].obs["pseudotime"],"pearson"))
r2 = [i*i for i in r]
Show code cell source
compare = pd.DataFrame({"bic":bic_res["bic.marginal"],"r2":r2})
spearman_corr = compare.corr(method="spearman").iloc[0,1]
# plot
plt.scatter(x=r2,y=bic_res["bic.marginal"])
plt.xlabel('R2 (truth vs pseudotime)')
plt.ylabel('scDesign3 BIC')
plt.text(x=0.05,y=450000,s="Spearman Correlation: %.2f" % spearman_corr)
plt.show()