Simulate datasets with multiple lineages
Introduction
In this example, we will show how to use scDesign3Py to simulate multiple lineages single-cell data.
Import packages and Read in data
import pacakges
import anndata as ad
import numpy as np
import scDesign3Py
Read in the reference data
The raw data is from the GEO with ID GSE72859, which describes myeloid progenitors from mouse bone marrow.
We pre-select the top 1000 highly variable genes. To save time, we only use the top 30 genes.
data = ad.read_h5ad("data/MARROW.h5ad")
data = data[:,0:30]
data
View of AnnData object with n_obs × n_vars = 2660 × 30
obs: 'Seq_batch_ID', 'Amp_batch_ID', 'well_coordinates', 'Plate_ID', 'Pool_barcode', 'Cell_barcode', 'CD34_measurement', 'FcgR3_measurement', 'cluster', 'Size_Factor', 'cell_type', 'Pseudotime', 'State', 'cell_type2', 'ery_meg_lineage_score', 'cell_stemness_score', 'no_expression', 'sizeFactor', 'pseudotime1', 'pseudotime2', 'l1', 'l2'
var: 'gene_short_name'
obsm: 'X_umap'
As we can see, this example dataset has two sets of pseudotime, thus two lineages. The variables pseudotime1 and pseudotime2 contain the corresponding pseudotime for each cell. The variables l1 and l2 indicate whether a particular cell belong to the first and/or second lineages.
data.obs[["pseudotime1","pseudotime2","l1","l2"]].head()
| pseudotime1 | pseudotime2 | l1 | l2 | |
|---|---|---|---|---|
| W31105 | 0.950862 | 0.568357 | TRUE | TRUE |
| W31106 | 9.168276 | -1.000000 | TRUE | FALSE |
| W31107 | -1.000000 | 7.981990 | FALSE | TRUE |
| W31108 | 11.394132 | -1.000000 | TRUE | FALSE |
| W31109 | -1.000000 | 8.080133 | FALSE | TRUE |
Simulation
Then, we can use this multiple-lineage dataset to generate new data by setting the parameter mu_formula as two smooth terms for each lineage.
test = scDesign3Py.scDesign3(n_cores=3,parallelization="pbmcmapply")
test.set_r_random_seed(123)
simu_res = test.scdesign3(
anndata=data,
default_assay_name="counts",
celltype="cell_type",
pseudotime=["pseudotime1", "pseudotime2", "l1", "l2"],
mu_formula="s(pseudotime1, k = 10, by = l1, bs = 'cr') + s(pseudotime2, k = 10, by = l2, bs = 'cr')",
sigma_formula="1",
family_use="nb",
usebam=False,
corr_formula="1",
copula="gaussian",
)
Then we can construct new data using the simulated count matrix.
simu_data = ad.AnnData(X=simu_res["new_count"], obs=simu_res["new_covariate"])
simu_data.layers["log_transformed"] = np.log1p(simu_data.X)
data.layers["log_transformed"] = np.log1p(data.X)
Visualization
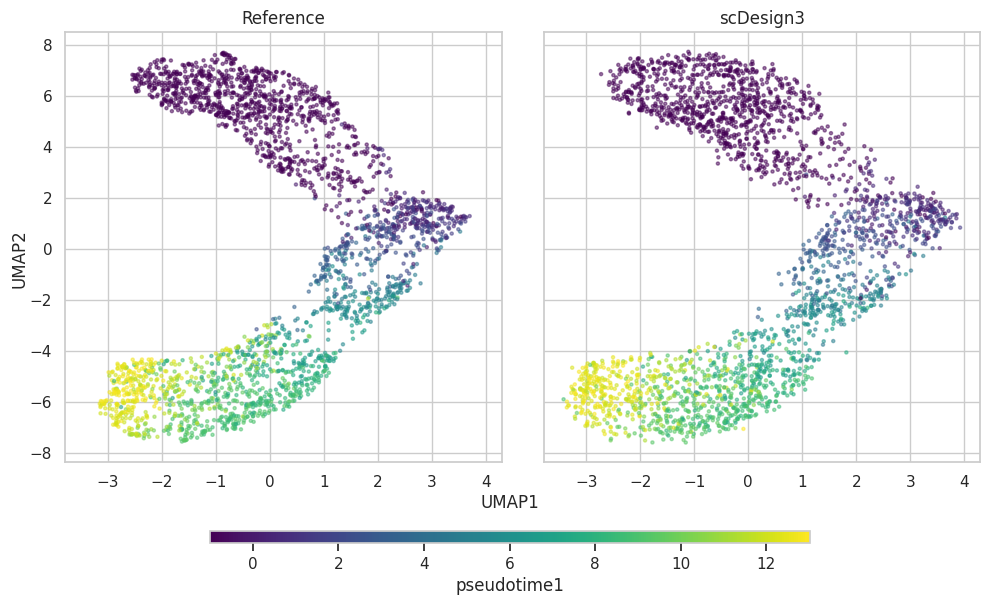
plot1 = scDesign3Py.plot_reduceddim(
ref_anndata=data,
anndata_list=simu_data,
name_list=["Reference", "scDesign3"],
assay_use="log_transformed",
if_plot=True,
color_by="pseudotime1",
n_pc=20,
point_size=5,
)
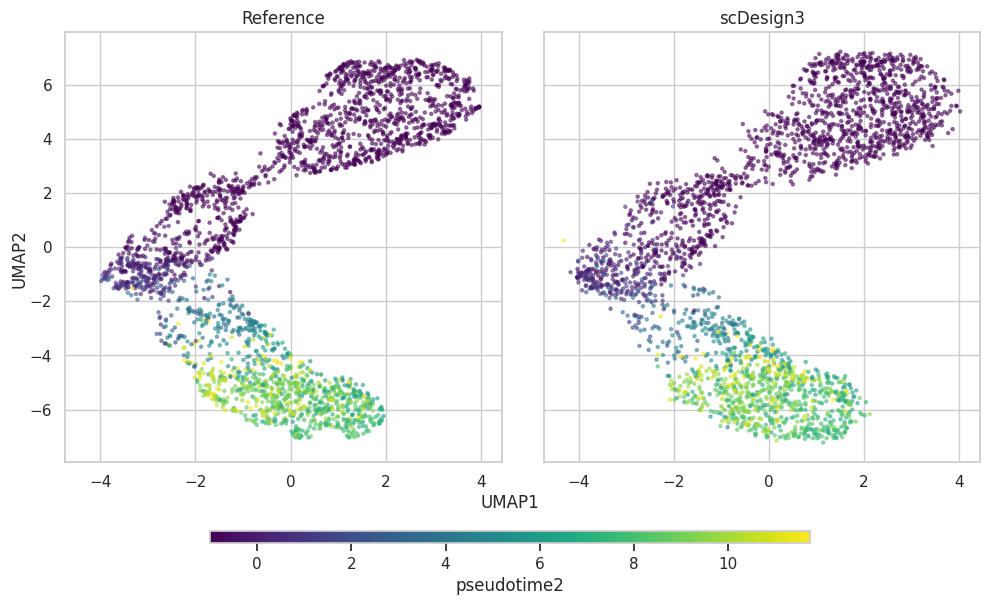
plot2 = scDesign3Py.plot_reduceddim(
ref_anndata=data,
anndata_list=simu_data,
name_list=["Reference", "scDesign3"],
assay_use="log_transformed",
if_plot=True,
color_by="pseudotime2",
n_pc=20,
point_size=5,
)
Pseudotime1
plot1["p_umap"]
Pseudotime2
plot2["p_umap"]